Motivation
Spectroscopy is a measurement technique that can be used to estimate compositions by observing the interaction of matter and electromagnetic (EM) radiation, such as light. In Raman spectroscopy, the unique spectral signature of a molecule is composed of a series of peaks that correspond to the energy levels of its chemical bonds. Sir Chandrashekhara V. Raman was awarded the Nobel Prize in Physics in 1930 for discovering this method of spectroscopy. More recently, with the advent of lasers and digital image sensors, applications of Raman spectroscopy have become widespread (Ellis et al. 2013; Noonan et al. 2018).

Figure 1: The mast of the Mars 2020 "Perseverance" rover, with the SuperCam instrument (Image credit: NASA)
Scheduled to launch in July 2020, the Mars rover "Perseverance" will be equipped with two Raman spectrometers, SuperCam and SHERLOC. This would be the first time that Raman spectroscopy has been performed on the Martian surface. The goal of this NASA mission will be to search for spectral signatures of biological molecules that might indicate the presence of life on another planet (Hutchinson et al. 2014). We are currently developing Bayesian methods for analysis of Raman spectroscopy, using pre-flight calibration data (Beyssac et al. 2020) as well as spectra from Mars-like environments on Earth, such as the CanMars Mars Sample Return Analogue Deployment (MSRAD) (Stromberg et al. 2019).
Let $\mathbf{Y}(\tilde\nu)$ be a vector of Raman scattering intensity values that have been discretised at a sequence of wavenumbers $\tilde\nu = (\nu_1, \nu_2, \dots, \nu_J)$. We are mainly concerned with wavenumbers in the range 800 to 1800 inverse centimetres $(\mathrm{cm}^{-1})$, which is the fingerprint region for organic molecules. In a Raman map, these intensity values can also have a spatial location $\mathbf{s}$ in a 2-D or 3-D coordinate system, as well as a time $t$. Then, $\mathbf{Y}(\mathbf{s}, t, \tilde\nu)$ forms a multi-dimensional data cube, which is sometimes referred to as a hyper-spectral observation in the remote sensing literature (Landgrebe, 1999).
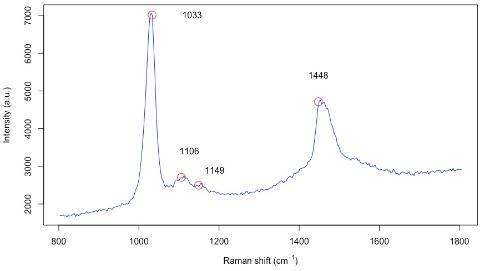
Figure 2: Observed Raman spectrum for methanol, with 4 characteristic peaks at 1033, 1106, 1149 and 1448 wavenumbers.